Benchmarking Neural F0 Estimation for Bioacoustics
Bioacoustic fundamental frequency estimation: a cross-species dataset and deep learning baseline
Paul Best, Marcelo Araya-Salas, Axel G. Ekström, Bárbara Freitas, Frants H. Jensen, Arik Kershenbaum, Adriano R. Lameira, Kenna D. S. Lehmann, Pavel Linhart, Robert C. Liu, Malavika Madhavan, Andrew Markham, Marie A. Roch, Holly Root-Gutteridge, Martin Šálek, Grace Smith-Vidaurre, Ariana Strandburg-Peshkin, Megan R. Warren, Matthew Wijers, Ricard Marxer
✉️ Contact authors: Paul Best paul.best@univ-amu.fr, Ricard Marxer ricard.marxer@lis-lab.fr
📄 Full paper (Author Accepted Manuscript):
Download PDF
🧠 Overview
The fundamental frequency (F0) is essential to characterising animal vocalisations, supporting studies from individual recognition to population dialects.
Yet:
- ⚠️ Manual annotation is time-consuming
- ⚙️ Automation is complex — especially in bioacoustics
🔍 Motivation
While speech and music research has made great strides in automatic F0 tracking, bioacoustics lags behind. Why?
- Extreme variation in signal properties (frequency, harmonicity, SNR)
- Presence of non-linear phenomena
📦 Our Contribution
To close this gap, we introduce:
- 📊 A benchmark dataset of over 250,000 vocalisations from 14 taxa, with ground truth F0
- 🐦 A diversity of signals: infra- to ultra-sound, high to low harmonicity
- 🤖 A benchmark of F0 estimation algorithms, including deep learning models
🧬 Key Findings
- ✅ Deep learning-based methods can:
- generalise to species not seen during training
- leverage unlabeled data via self-supervision
-
🧠 Self-supervised models can perform nearly as well as supervised ones
- 📈 We propose spectral quality metrics that correlate with F0 tracking performance
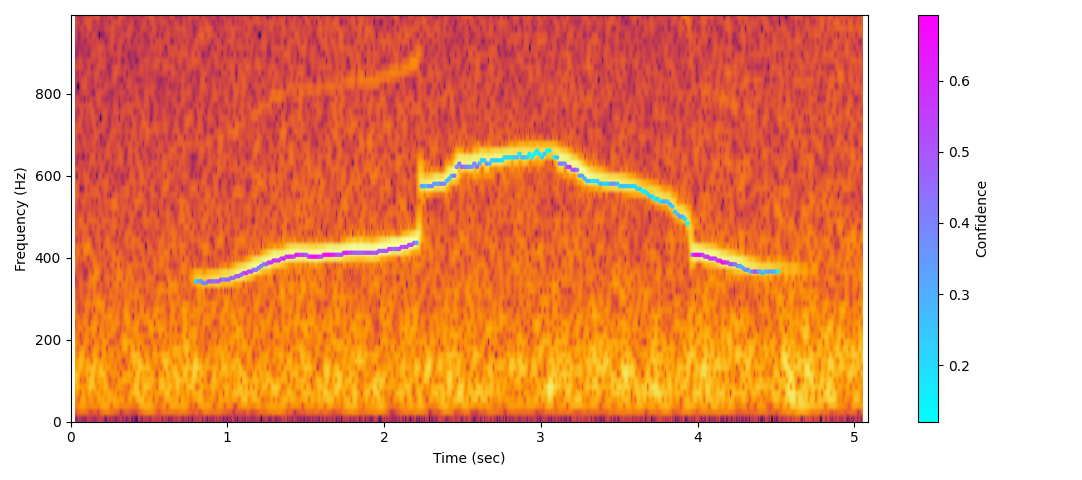
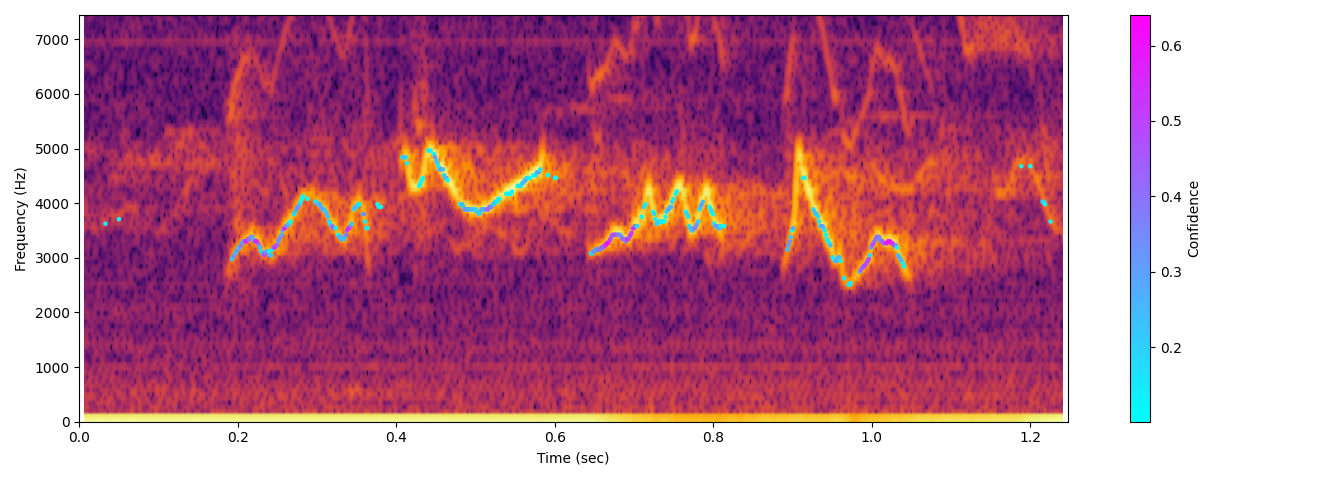
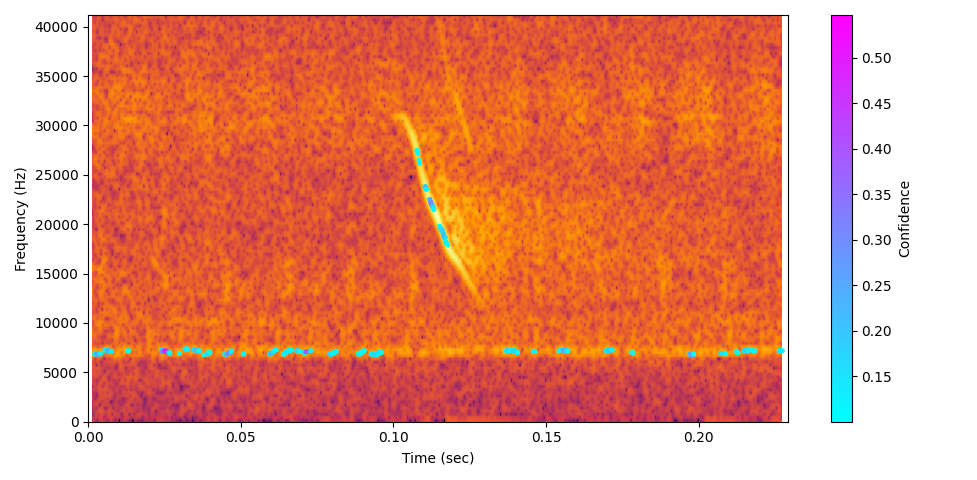
🔊 Example Visualisations & Audio
Wolf howl
Réunion grey white-eye (slowed x5)
Bat echolocation (slowed x20)
🛠️ Resources
-
💻 Code & Pretrained Models:
GitHub – mim-team/bioacoustic_F0_estimation
Includes an easy-to-use command-line script for F0 estimation:python predict.py my_sound_file.wavIn addition to F0, the tool also extracts several spectral features:
- 🎯 Sub-harmonic ratio
- 🔊 Harmonicity
- 📈 F0 salience
These descriptors help assess signal quality and are useful for downstream analysis.
🌍 Broader Impact
This study is the first large-scale adaptation of Music Information Retrieval (MIR) tools to the rich, complex world of bioacoustic signals.
While no model is perfect, our work shows that deep learning offers a promising path toward generic, scalable, and reliable F0 estimation in bioacoustics.
The estimated F0 contours, together with additional features like harmonicity and F0 salience, can support interpretable vocalisation categorisation — whether for individual signatures, repertoire categories, or other groupings. This provides a valuable middle ground between manual expert annotation and black-box machine learning, facilitating both scientific insight and scalable analysis.
🔗 Read the full article:
“Bioacoustic fundamental frequency estimation: a cross-species dataset and deep learning baseline”
Published in Bioacoustics: The International Journal of Animal Sound and its Recording (2025)
📚 How to Cite
If you use this work in your research, please cite:
@article{Best02062025,
author = {Paul Best and Marcelo Araya-Salas and Axel G. Ekström and Bárbara Freitas and Frants H. Jensen and Arik Kershenbaum and Adriano R. Lameira and Kenna D. S. Lehmann and Pavel Linhart and Robert C. Liu and Malavika Madhavan and Andrew Markham and Marie A. Roch and Holly Root-Gutteridge and Martin Šálek and Grace Smith-Vidaurre and Ariana Strandburg-Peshkin and Megan R. Warren and Matthew Wijers and Ricard Marxer},
title = {Bioacoustic fundamental frequency estimation: a cross-species dataset and deep learning baseline},
journal = {Bioacoustics},
volume = {0},
number = {0},
pages = {1--28},
year = {2025},
publisher = {Taylor & Francis},
doi = {10.1080/09524622.2025.2500380},
URL = {https://doi.org/10.1080/09524622.2025.2500380},
eprint = {https://doi.org/10.1080/09524622.2025.2500380}
}